$ 0.013 0.80%
Hdac (HDAC) Rank 9345
| Mkt.Cap | $ 0.00000000 | Volume 24H | 0.00000000HDAC |
| Market share | 0% | Total Supply | 0.00000000HDAC |
| Proof type | ePoW | Open | $ 0.01 |
| Low | $ 0.01 | High | $ 0.01 |
Histone Modifications
In other words, although hyperacetylated histones interact with transcription activators, HDACs may furnish deacetylated interaction sites for transcriptional repressors. More recently, using high-resolution mass spectrometry, more than 3600 acetylation sites on 1750 proteins were identified (Choudhary et al. 2009). The acetylation sites are present on nuclear, cytoplasmic, and mitochondrial proteins involved in many different cellular processes. The use of suberoylanilide hydroxamic acid (SAHA) and MS-275, two broad spectrum Class I, II, IV HDAC inhibitors, up-regulated ∼10% of all acetylation sites by at least a factor of 2, suggesting that many of these acetylations are regulated by classical HDACs.
How does Trichostatin a work?
In humans, there are 18 HDAC enzymes divided into four classes: the Class I Rpd3-like proteins (HDAC1, HDAC2, HDAC3, and HDAC8); the Class II Hda1-like proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10); the Class III Sir2-like proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7); and the Class IV
DNA Methylation
How does methylation work?
Their function is to package DNA into structural units called nucleosomes. Histones are the main proteins in chromatin. Chromatin is a combination of DNA and protein which makes up the contents of a cell nucleus. Because DNA wraps around histones, they also play a role in gene regulation.
Histone modifications act in diverse biological processes such as transcriptional activation/inactivation, chromosome packaging, and DNA damage/repair. Transcription is ultimately regulated by the interaction of multiple epigenetic mechanisms that cooperate to activate or silence gene expression. Methylation is regulated by proteins such as Dnmt and Tet (purple) that are involved in the active addition or chemical modification (such as hydroxymethylation in red) of DNA methylation. To suppress gene expression, Dnmts target CpG sites and actively methylate DNA. For some Dnmts, their catalytic activity is enhanced by association with histone tails and with Dnmt3L.

The lysine-specific transferases are further broken down into whether or not they have a SET domain or a non-SET domain. These domains specify exactly how the enzyme catalyzes the transfer of the methyl from SAM to the transfer protein and further to the histone residue.[9] The methyltransferases can add 1-3 methyls on the target residues.

Where does DNA methylation occur?
DNA methylation regulates gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factor(s) to DNA. During development, the pattern of DNA methylation in the genome changes as a result of a dynamic process involving both de novo DNA methylation and demethylation.
HDAC6 is a member of class II, which is subgrouped into class IIa (HDAC4, 5, 7, and 9) and class IIb (HDAC6 and 10). Class II HDACs shuttle between the nucleus and the cytoplasm, with the exception of HDAC6, which is mainly localized in the cytosol. Other researchers have used bacterial DNA-binding proteins to research Salmonella enterica serovar Typhimurium, in which the T6SS genes are activated from a macrophage infection.
Such alterations include point mutations, deletions, insertions, and translocation. In contrast, epigenetics is the study of heritable changes in gene activity or function that is not associated with any change of the DNA sequence itself.
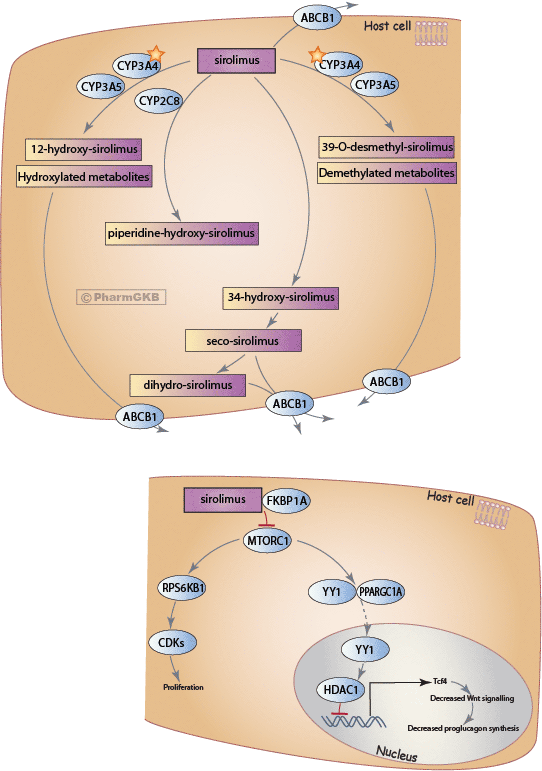
Histone acetylation and deacetylation are essential parts of gene regulation. These reactions are typically catalysed by enzymes with "histone acetyltransferase" (HAT) or "histone deacetylase" (HDAC) activity. Acetylation is the process where an acetyl functional group is transferred from one molecule (in this case, Acetyl-Coenzyme A) to another. Deacetylation is simply the reverse reaction where an acetyl group is removed from a molecule.
It has been found that H-NS and RNA polymerase both bind to the P1 promoter and form a complex. When H-NS is bound with RNA Polymerase to the promoter region, there are structural differences in the DNA that are accessible.[10] It has also been found that H-NS can affect translation as well by binding to mRNA and causing its degradation.
They contribute to the erasure of epigenetic modifications, help establish epigenetic off chromatin states, and regulate heritable changes in gene expression. How HDACs achieve these ends is a question of immediate interest to many working in this field. The finding that HDACs function as transcriptional repressors and corepressors has ushered a surge of interest in this subject, and reports of genes that are regulated by HDACs are continuously expanding. Global expression profiling experiments estimate that the transcription of 10% of genes are regulated by HDACs. However, questions of how each HDAC uniquely regulates a specific set of genes remain largely unexplored.
Similarly, although we know that there are potentially more than a thousand HDAC substrates, the biological consequences of deacetylation of these substrates require intense research. The mechanistic details of how histone deacetylation promotes modification cross talk, and the global consequences of deacetylation of other histone and epigenetic-modifying enzymes are ripe for further research.
It did this by decreasing the number through increasing the size of aggregates in a cellular model of Parkinson’s disease. Thus, development of small molecules modulators of sirtuin activity has become one of the most active areas in drug discovery. Sirtinol and splitomicin are identified as the first small molecule sirtuin inhibitors that affect telomere silencing in yeast.
What is a nucleosome made up of?
Explain how the acetylation of core histones may loosen chromatin packing. Acetylation can be defined as the process of the transfer of an acetyl group from acetyl-coenzyme A to any other molecule. DNA is held within the histone protein by the positive charge on lysine, which is present in histones.
Euchromatin and Heterochromatin

- Hence, when methylation is removed as a result of neuronal activity, it is not surprising that MBDs are often released from promoters (Martinowich et al, 2003).
- The mice survive for 3 -4 months, at which point they show massive cardiac hypertrophy and depression of the genes that control fatty-acid uptake and metabolism [36].
- In mdx hearts, global elevation of histone acetylase activity is associated with lateralized Cx43 and increased physical interaction between the acetylase P300/CBP-associated factor with Nε-lysine acetylated Cx43.
- The intracisternal A particle (IAP) is one of most aggressive retroviruses in the mouse genome (Walsh et al, 1998).
Hence, although both DNA methylation and demethylation are altered by neuronal activity, DNA methylation functions alongside other regulatory proteins and epigenetic mechanisms that determine gene expression. Interactions between nucleosomes allow for higher-order structures to form. These higher-order structures can condense the chromatin to the point where chromosomes form. In most species, histone H3 is primarily acetylated at lysines 9, 14, 18, 23, and 56, methylated at arginine 2 and lysines 4, 9, 27, 36, and 79, and phosphorylated at ser10, ser28, Thr3, and Thr11.
Advances in Molecular Toxicology
The Class I proteins (HDAC1, HDAC2, HDAC3, and HDAC8) have sequence similarity to the yeast Rpd3 protein. The Class II proteins (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10) have sequence similarity to the yeast Hda1 protein. Three proteins in Saccharomyces cerevisiae—Hos1, Hos2, and Hos3—however, have 35%–49% identity to Rpd3, and 21%–28% identity to Hda1. Thus, mammalian Class I and II HDACs are also related to the yeast Hos proteins. The Class III proteins (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7) have sequence similarity to the yeast Sir2 protein.
Role in DNA replication
It is not clear what structural implications histone phosphorylation has, but histone phosphorylation has clear functions as a post-translational modification, and binding domains such as BRCT have been characterised. What was said above of the chemistry of lysine methylation also applies to arginine methylation, and some protein domains—e.g., Tudor domains—can be specific for methyl arginine instead of methyl lysine. Arginine is known to be mono- or di-methylated, and methylation can be symmetric or asymmetric, potentially with different meanings. The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to their water solubility.
What effect would inhibitors of histone deacetylases have upon transcription?
Histone deacetylase (HDAC) inhibitors are a relatively new class of anti-cancer agents that play important roles in epigenetic or non-epigenetic regulation, inducing death, apoptosis, and cell cycle arrest in cancer cells.
Also, activation or localization of two histone acetyltransferases can be oncogenic. As mentioned above the histone tails have been shown to directly interact with the DNA of the nucleosome. Each histone in the octamer has an N-terminal tail that protrudes from the histone core. The tails play roles both in inter and intra nucleosomal interactions that ultimately influence gene access.[12] Histones are positively charged molecules which allow a tighter bonding to the negatively charged DNA molecule. The parts of the tail closest to the DNA hydrogen bond and strengthen the DNA's association with the octamer; the parts of the tail furthest away from the DNA, however, work in a very different manner.

Proper coordination of cell signaling with histone/non-histone deacetylation is critical to precisely regulate both gene expression and a number of transcription independent events. HDAC proteins are vital regulators of fundamental cellular events, such as cell cycle progression, differentiation, and tumorigenesis. Abnormal HDACs can contribute to many different human diseases including cancer, neurodegenerative disorders, cardiac hypertrophy, and pulmonary diseases.
(C) Phosphorylation of a Class II HDAC, HDAC4, promotes its interaction with the protein and, subsequently, changes its localization. Multiple residues (S246, S467, and S632 on HDAC4, and corresponding conserved sites on HDAC5, HDAC7, and HDAC9) confer the HDAC– interactions. HDAC11 regulates the protein stability of DNA replication factor CDT1 (Glozak and Seto 2009) and the expression of interleukin 10.
What are the two basic functions of histones?
In human DNA, 5-methylcytosine is found in approximately 1.5% of genomic DNA. In somatic cells, 5-mC occurs almost exclusively in the context of paired symmetrical methylation of a CpG site, in which a cytosine nucleotide is located next to a guanidine nucleotide.
Hypertrophic stimuli induced the expression of heat shock protein (Hsp) 70. Hsp70 overexpression produced a hypertrophic phenotype, which was blocked either by siHDAC2 or by a dominant negative Hsp70DeltaABD. These results suggest that the induction of Hsp70 in response to diverse hypertrophic stresses and the ensuing activation of HDAC2 trigger cardiac hypertrophy.
HDAC2 is similarly phosphorylated at residues S394, S422, and S424 (corresponding to S393, S421, and S423 of HDAC1; Tsai and Seto 2002). Its phosphorylation also promotes enzymatic activity and affects protein complex formation with Sin3 and Mi2.
In regions of DNA with activate transcription, Tet removes DNA methylation, and histone tails in this region often contain H3K4me3 that inhibits Dnmt binding to unmethylated CpG sites and maintains a permissive environment for transcription. TSA can be used to alter gene expression by interfering with the removal of acetyl groups from histones (histone deacetylases, HDAC) and therefore altering the ability of DNA transcription factors to access the DNA molecules inside chromatin. It is a member of a larger class of histone deacetylase inhibitors (HDIs or HDACIs) that have a broad spectrum of epigenetic activities.

One possibility is that HDACs may down-regulate transcription of transcriptional repressors, which leads to derepression of gene expression. Alternatively, HDACs may deacetylate and, consequently, activate transcription activators or inhibit the functions of transcription repressors independent of histone modifications. In summary, there is overwhelming evidence that a key biological function of HDACs is to modulate transcription, especially in repression. However, whether HDACs can directly activate transcription and the exact detailed mechanisms by which they regulate transcription still remain to be determined.
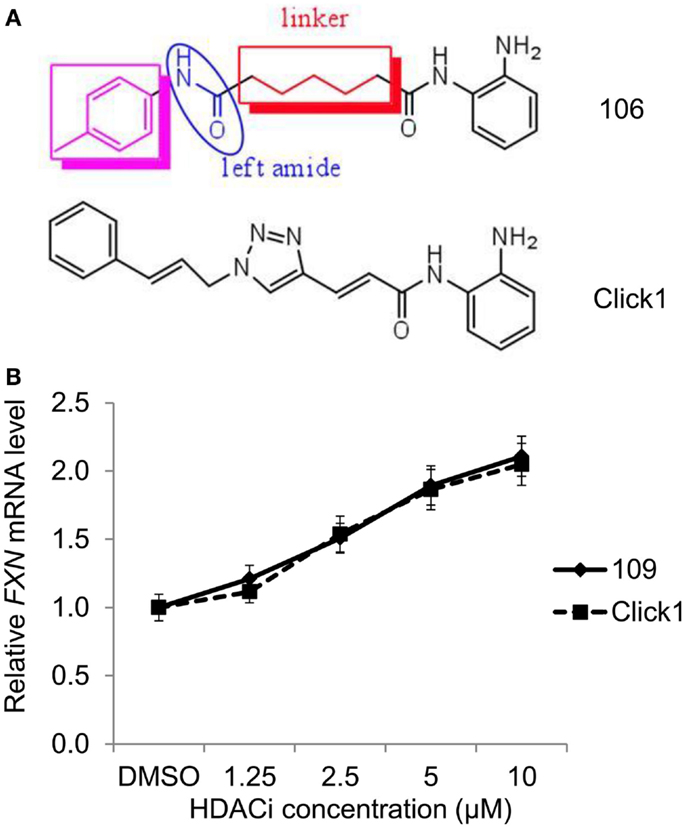
Malfunction of SIRT1 may contribute to diabetes, obesity, abnormal cancer metabolism, cancer stemness, and neurological disorders. These diseases represent a huge burden to our societies’ health and healthcare systems, therefore sirtuin inhibitors represent hopeful therapeutics for many of these diseases.







